Genome browser
Attention: You are viewing a page from the old CRC user manual.
This page may not represent the most up to date version of the content.
You can view it in our new user manual here.
IGV
The Integrative Genomics Viewer (IGV) is a high-performance, easy-to-use, interactive tool for the visual exploration of genomic data. Tthe original IGV is a Java desktop application. You can run IGV as a VNC application through Open Ondemand.
Logon ondemand.htc.crc.pitt.edu. Click Interactive Apps -> IGV on htc:

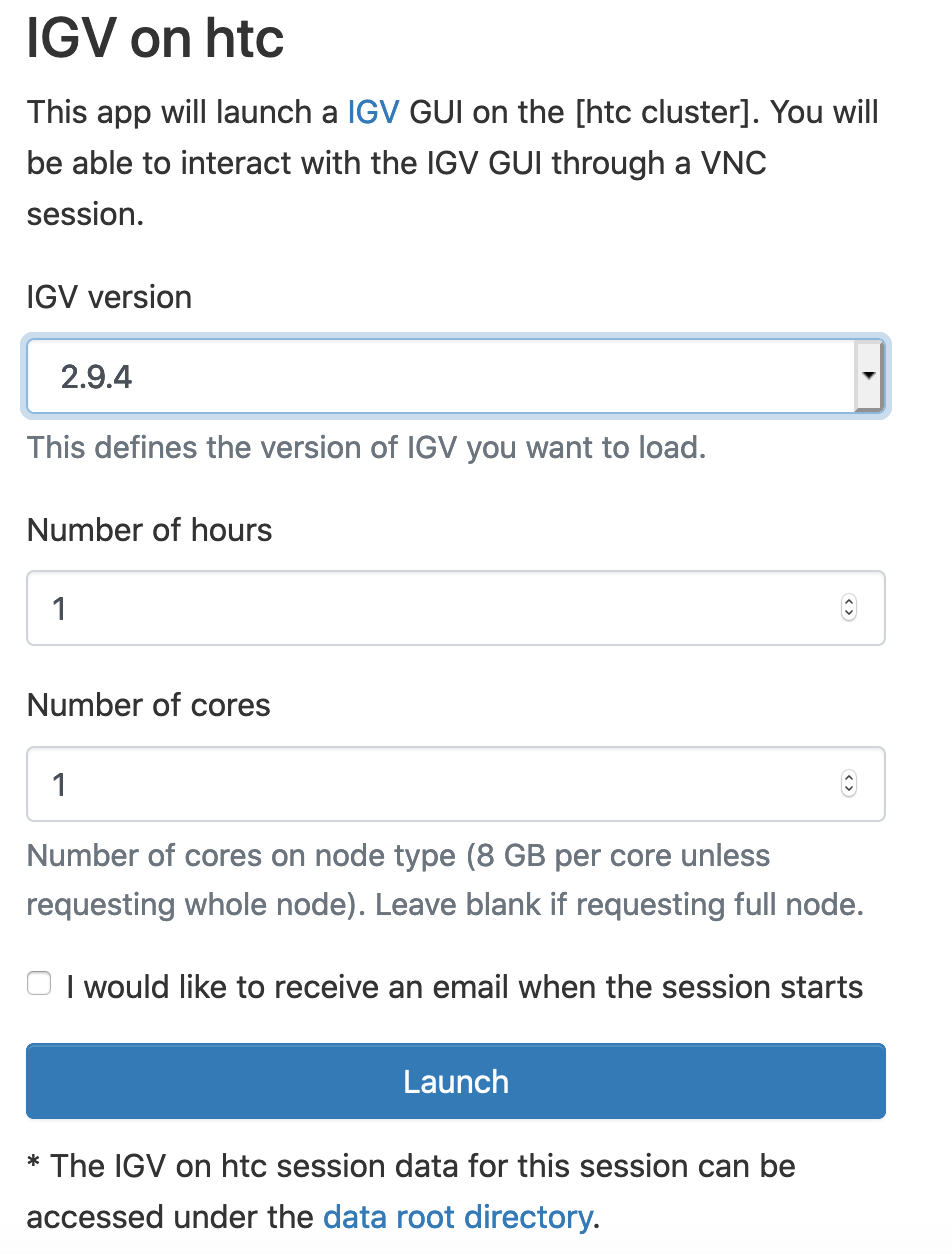
Select IGV version, Number of Hours and Number of cores. Click Launch:
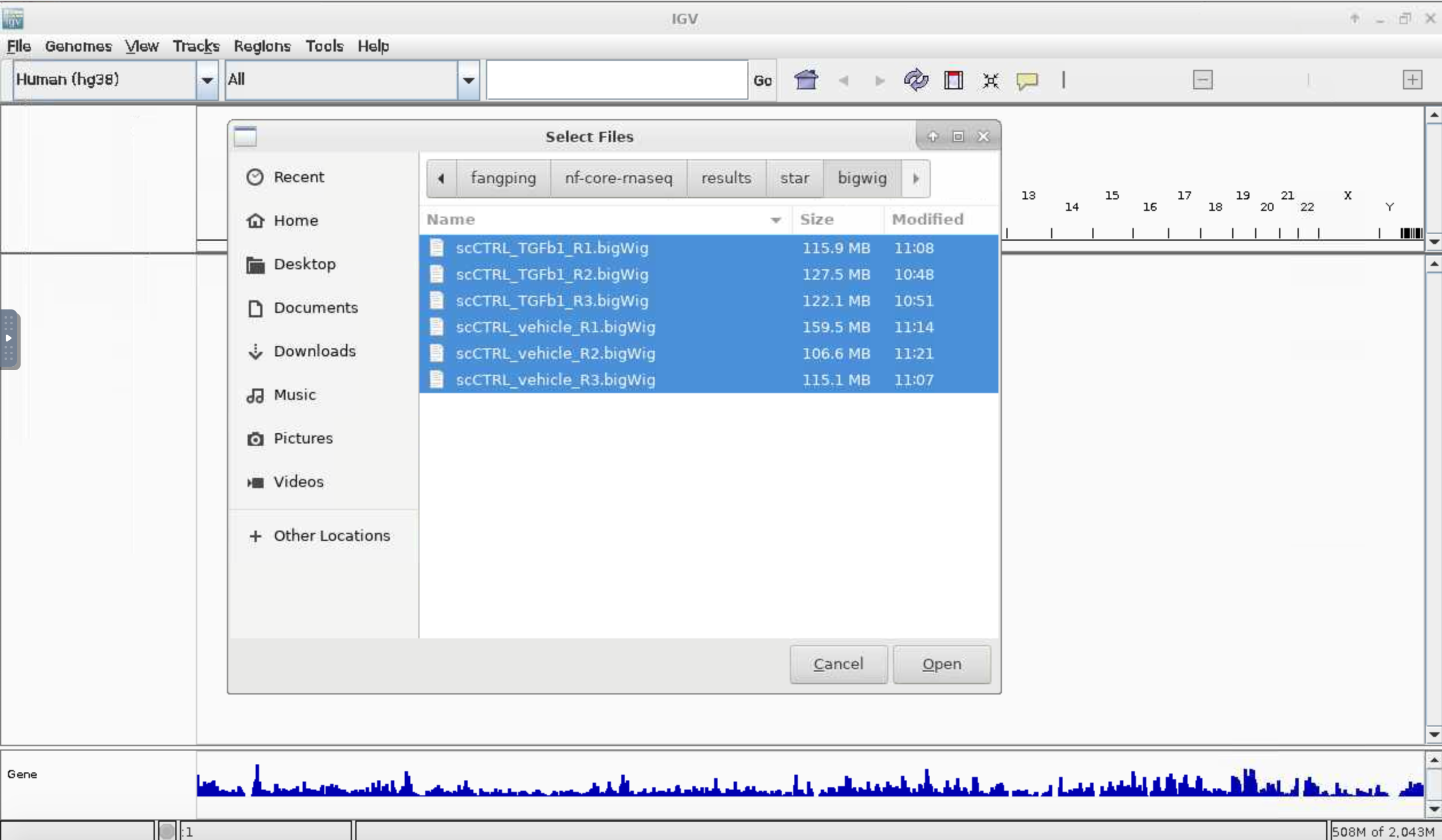
Choose your genome at the top left cornet. I have selected Human (hg38). File -> Load from File... and navigate to your CRC folder. In the screenshot, I have navigated to /bgfs/genomics/fangping/nf-core-rnaseq/results/star/bigwig, and selected 6 bigWig files.
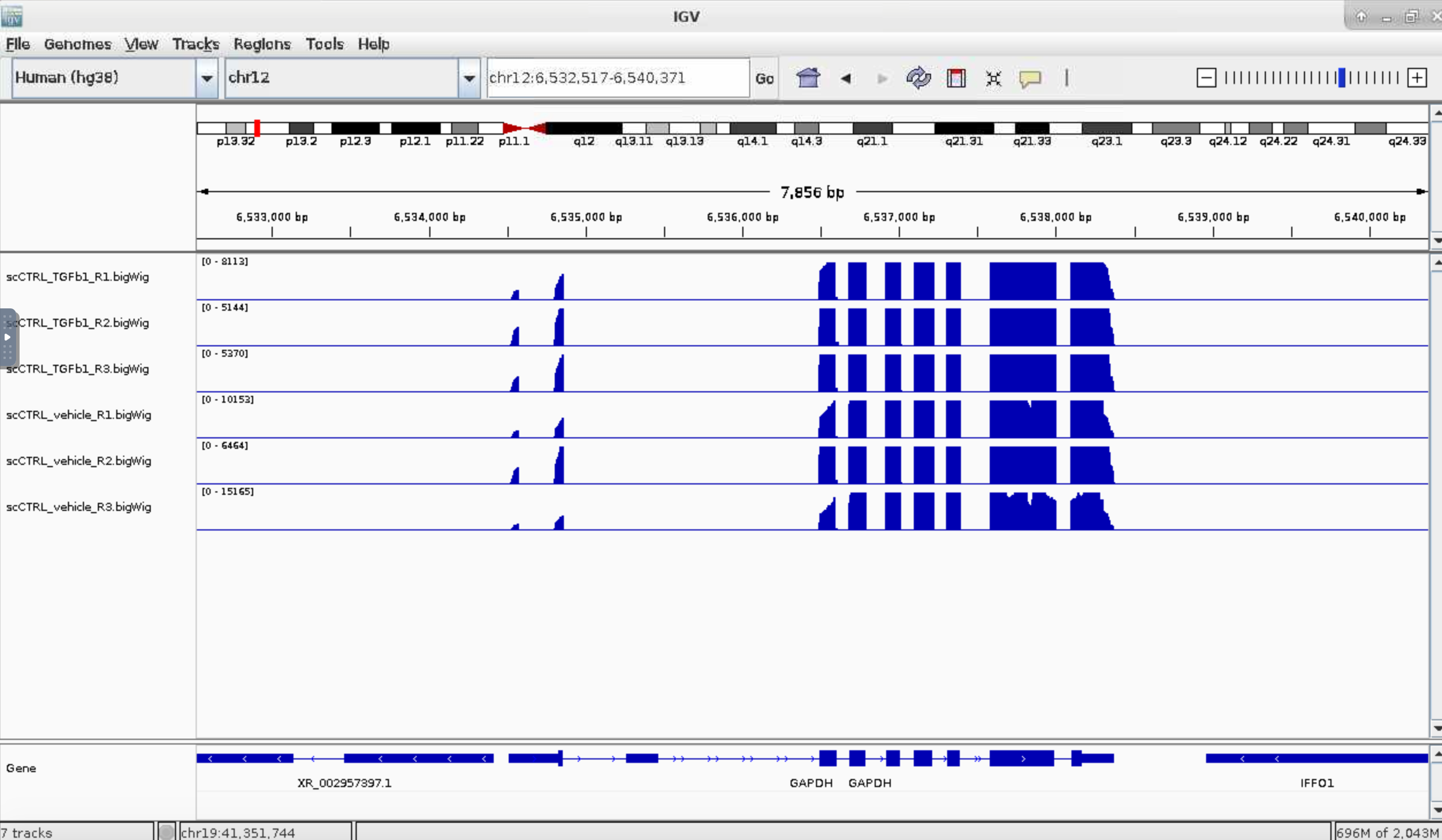
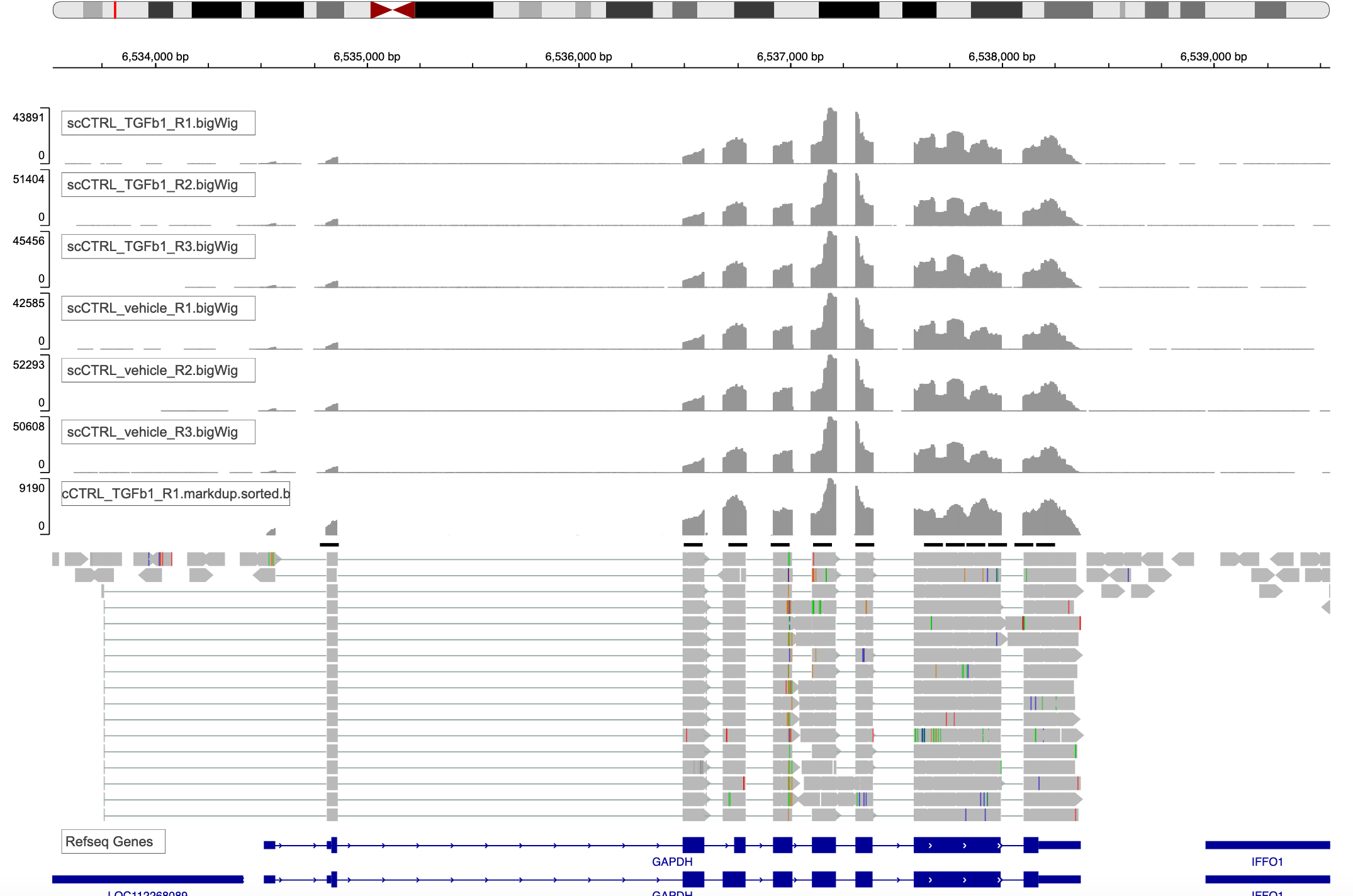
Type a gene name in top middle text field. I have used GAPDH.
IGV_webapp
The IGV-Web app is a pure-client "genome browser" application based on igv.js. IGV-web is a web application.
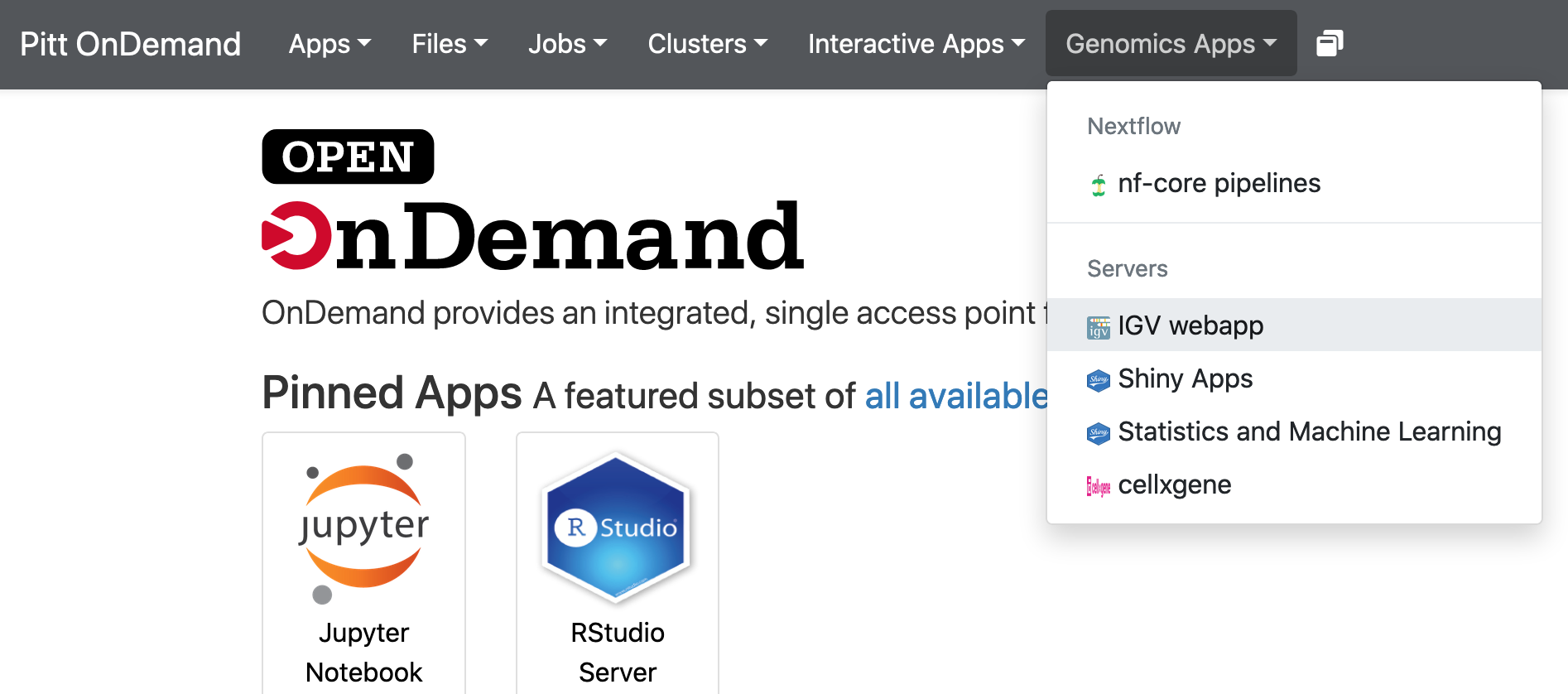
Logon ondemand.htc.crc.pitt.edu. Click Genomics Apps -> IGV webapp:
Select IGV webapp version, Number of cores and Number of hours. Click Launch:
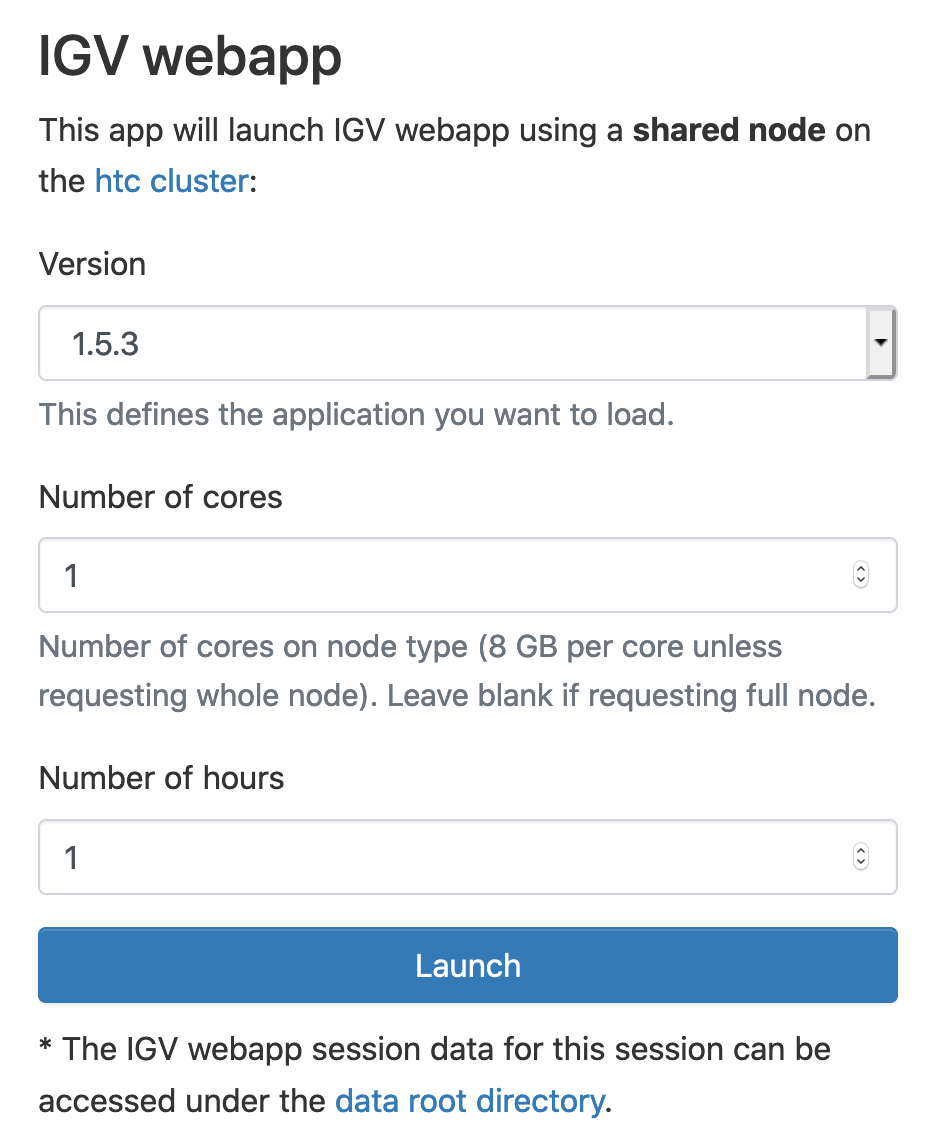
This will launch IGV-webapp using a shared node on the htc cluster.
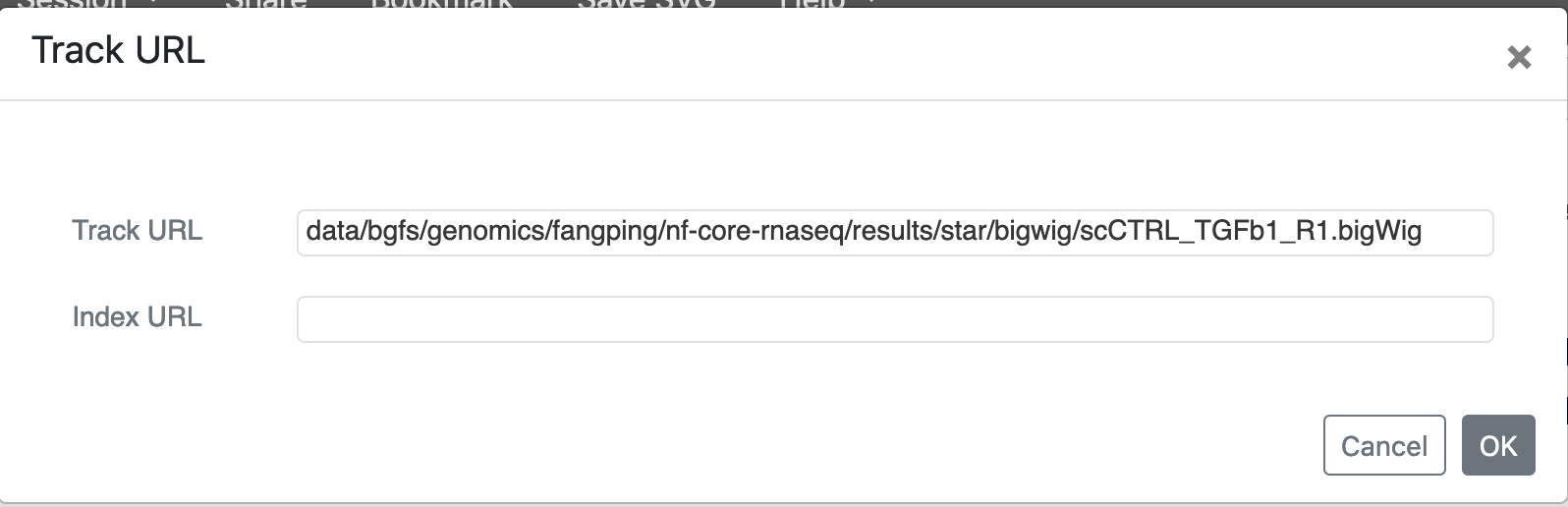
Choose your genome at the top left corner. To add files on CRC storage as track, select Tracks -> URL ... Get the absolute path of your file, for example,
/bgfs/genomics/fangping/nf-core-rnaseq/results/star/bigwig/scCTRL_TGFb1_R1.bigWig,
then add data in front. Use that address, i.e.,
data/bgfs/genomics/fangping/nf-core-rnaseq/results/star/bigwig/scCTRL_TGFb1_R1.bigWig,
as Track URL.
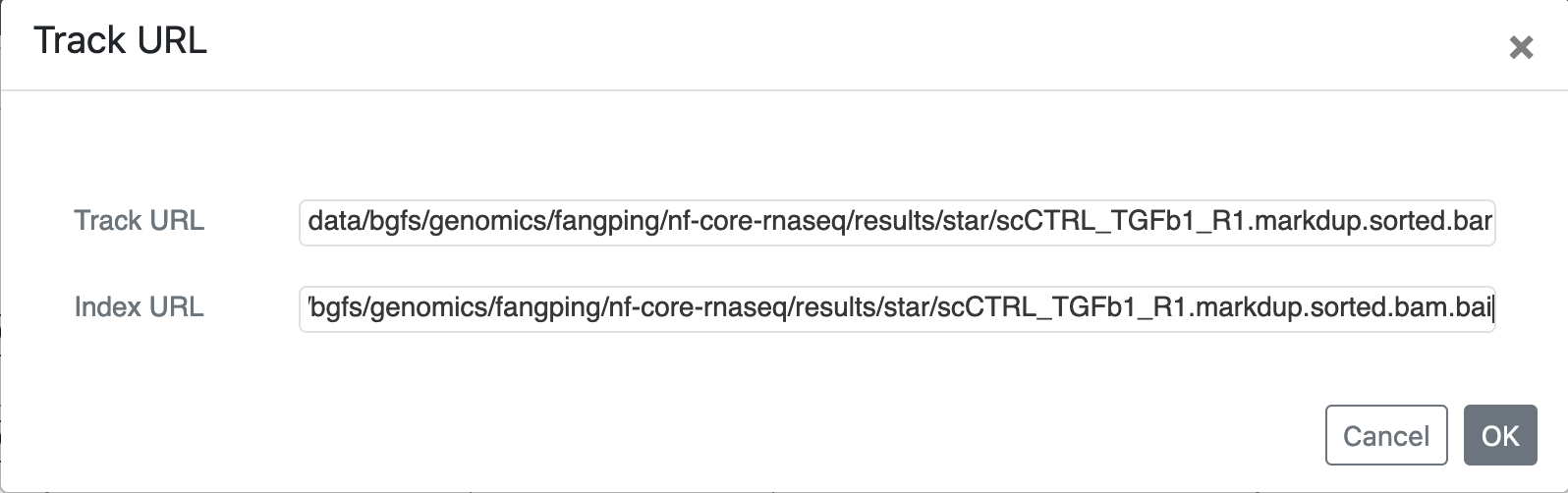
When track has index, you should provide Index URL. For example, bam file /bgfs/genomics/fangping/nf-core-rnaseq/results/star/scCTRL_TGFb1_R1.markdup.sorted.bam and its index file /bgfs/genomics/fangping/nf-core-rnaseq/results/star/scCTRL_TGFb1_R1.markdup.sorted.bam.bai. You should use:
Track URL: data/bgfs/genomics/fangping/nf-core-rnaseq/results/star/scCTRL_TGFb1_R1.markdup.sorted.bam
Index URL: data/bgfs/genomics/fangping/nf-core-rnaseq/results/star/scCTRL_TGFb1_R1.markdup.sorted.bam.bai
Type a gene name in top middle text field. I have used GAPDH.
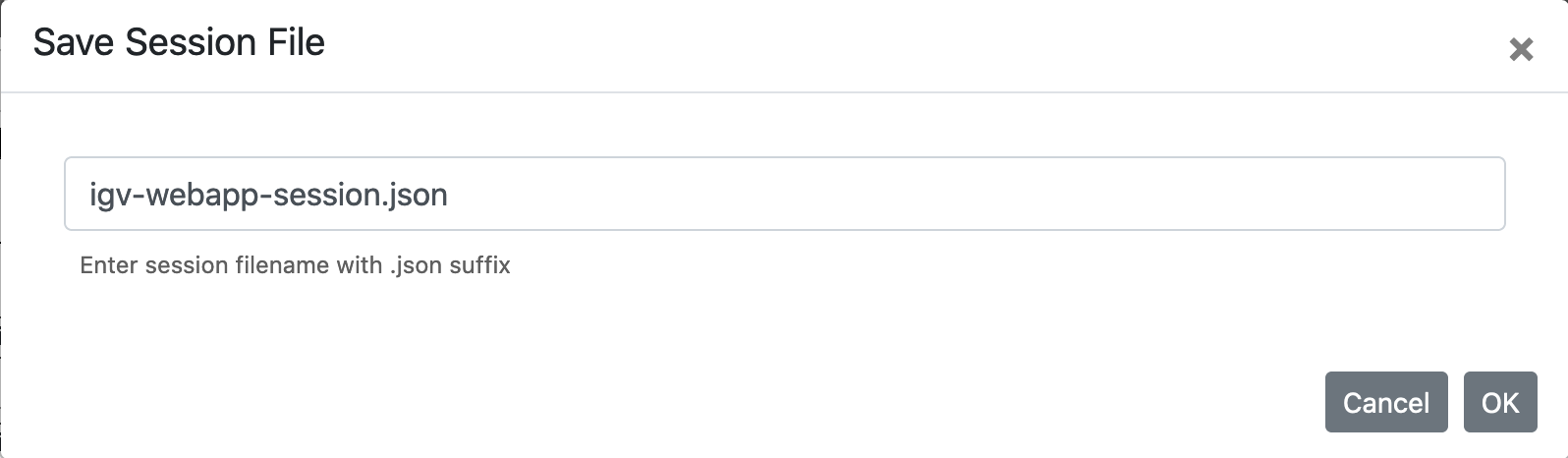
To save and share the session, choose Session -> Save ...., and save the json file to your local computer.
You can send this json file to your collaborator to reproduce the IGV session. Choose Session -> Local File ... to load the session.
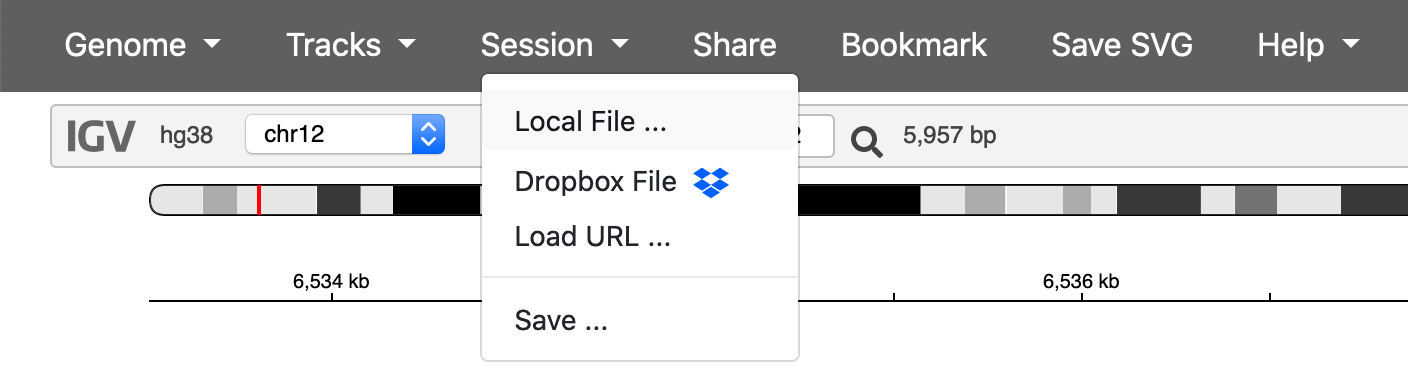
You can upload this file to generate the above session.
