Pitt researchers hunt viruses in Project Premonition

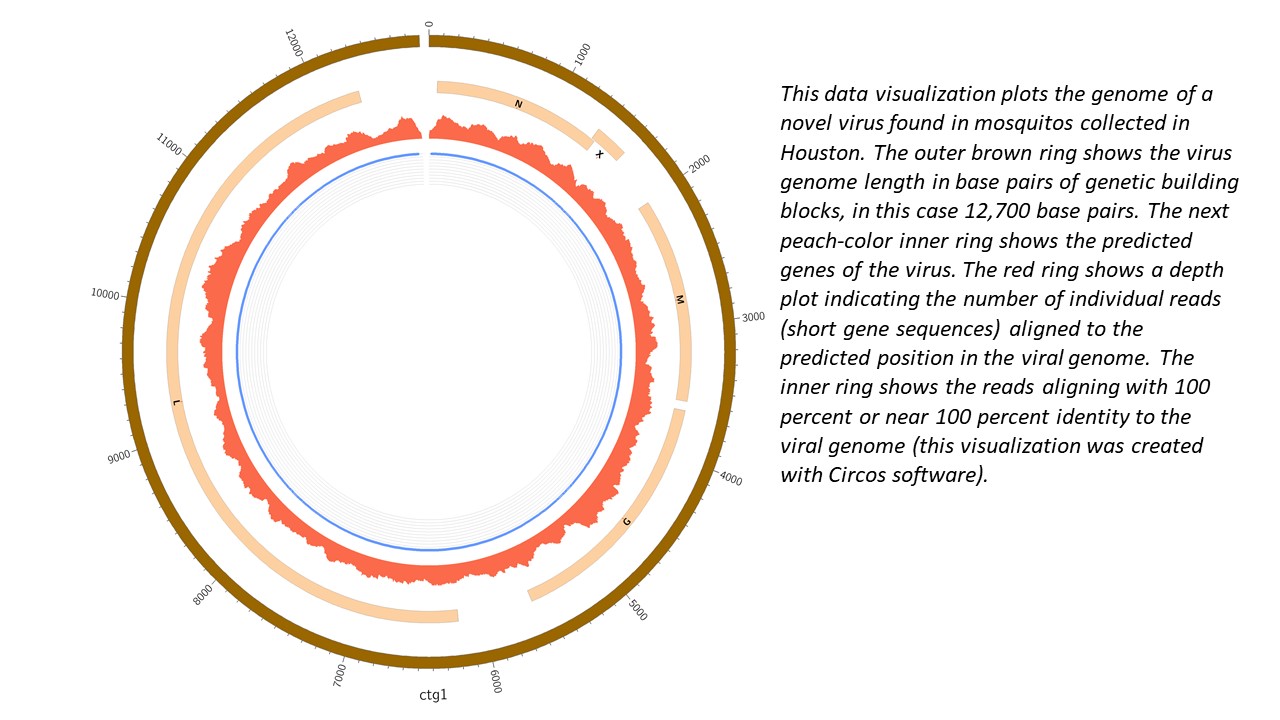
Pittsburgh prides itself as city of bridges, city of champions, Amazon HQ2 candidate and more, but Pittsburgh’s viral distinction is not widely known – the city is namesake of the Pittsburgh Sewage-Associated Virus 1 (PSAV1), initially described by Pitt biologist James Pipas in his 2011 paper “Raw Sewage Harbors Diverse Viral Populations.” The virus received the Pittsburgh moniker with the publication of its complete genome in 2018.
Why raw sewage? Pipas paraphrases bank robber Willie Sutton – “That’s where the money is. Sewage water is rich in viruses.” Pipas, professor and Herbert W. and Grace Boyer Chair in molecular biology, published the complete PSAV1 genome sequence in 2018, along with Paul Cantalupo, biology systems manager.
The raw sewage virome publication produced unexpected consequences when Microsoft Research scientists read the paper and invited the Pipas lab to join an ambitious virus-hunting effort called Project Premonition.
Project Premonition is a global strategy to predict outbreaks of mosquito-borne viruses, both existing and emerging. The Pipas lab’s role is sequencing and analyzing mosquito meta-genomes – genomes not only of mosquitoes, but also of the bloodmeal of humans, dogs and other creatures upon which the mosquitoes feed. Mosquitoes are located, captured, and identified using surveillance drones and robotic traps that have been tested in Harris County, Texas, the island of Grenada, and soon at the Jane Goodall Institute’s Gombe Stream Research Centre in Tanzania.
 In Texas alone, the traps collected 22,000 samples among nine species of mosquitoes, including species that carry Malaria and the Zika, Dengue, and West Nile viruses. The Pipas lab collaborated with Microsoft Research and Johns Hopkins University in developing a database of more than 250,000 genomes to match against the samples.
In Texas alone, the traps collected 22,000 samples among nine species of mosquitoes, including species that carry Malaria and the Zika, Dengue, and West Nile viruses. The Pipas lab collaborated with Microsoft Research and Johns Hopkins University in developing a database of more than 250,000 genomes to match against the samples.
The lab relies heavily on Pitt Center for Research Computing consultants and resources to analyze millions of gene sequences that may vary in length by multiple orders of magnitude, depending on the species. For each month in 2018, the Pipas lab used around 65,000 core hours, or the equivalent of running over 100 laptops flat out for the month.
Cantalupo is primarily responsible for the computation, in collaboration with Fangping Mu, Pitt CRC research assistant professor and lead consultant for this project. “Bioinformatics analyses involve transferring files in parallel through a series of steps, called a pipeline or a workflow,” Mu explains. “Typically, these transformations are done by existing command line software. But very often software packages are not compatible. Pitt CRC installs and configures the analysis packages, but I also spend a lot of time debugging.” Cantalupo describes Mu’s role as essential. “Fangping can install anything and make it work.”
Next generation sequencing (NGS) and subsequent analyses are often accurate in identifying viruses – but with a caveat. If a sample is contaminated, the virus hunt can lead down a rabbit hole. Beyond identifying viruses, Cantalupo and Pipas have experience finding mis-identified viruses. They have detected sequencing contamination in tumor samples that led to the mis-identification of Human Papillomavirus and Dengue.
Cantalupo says. “Jim has years of experience, so he often senses when a sequence doesn’t look right. But you can’t code intuition. Hunting for contamination is a significant time-sink, but we need to do it. Otherwise, what evidence do we really have? If we miss – or mis-identify – a virus, we defeat the whole purpose.”
Contact:
Brian Connelly
Coordinator of Outreach and Communications
bgc14@pitt.edu
